Research focus and objectives
TOP
Alzheimer's disease: new mechanistic insights for future therapies
Alzheimer's disease: new mechanistic insights for future therapies AD
Alzheimer's disease is the consequence of the accumulation in the brain of three lesions: amyloid-β (Aβ) and tau lesions and neuroinflammation. Our group evaluates mechanisms associated with the occurrence of these lesions and with the impact of these lesions on synaptic loss and functional impairments associated with Alzheimer's disease.
1.
Alzheimer's disease pathology is transmissible including in primates
AD1
Our group has demonstrated that Aβ and tau lesions are transmissible and can spread in the brain of mice and in primates (Gary, 2019). This transmission makes it possible to explore the pathophysiogenic mechanisms leading to the pathology.
In addition to studies in mice, we investigate Alzheimer pathology in primates (Microcebus murinus). The microcebe is a model of neurodegenerative pathologies linked to aging. This animal presents, as it ages, cognitive alterations, alterations in cerebral metabolism, cerebral atrophy and amyloid deposits.
Click on the picture to enlarge
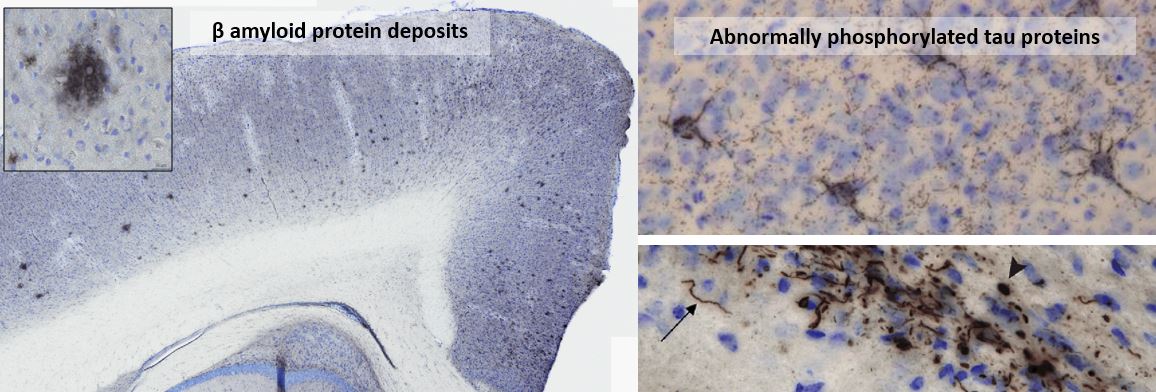
Amyloid and tau lesions induced in the brain of experimental models following inoculation of amyloid
and tau nucleating factors.
Examples of ongoing projects
- Assessment of the impact of purified forms of Aβ with very specific mutations (PI. M Dhenain, AS Herard, Vaincre Alzheimer, France Alzheimer, collaboration with Alain Buisson from Grenoble Institute of Neurosciences).
- Assessment of the impact of Aβ and tau lesions transmission on neuroinflammation, synaptic loss, and cerebral function (PI. M Dhenain, InductAlz, PrimAlz, collaboration with Stephane Haik (ICM, Paris), Luc Buée (Lille University)).
2.
Structural diversity of amyloids in Alzheimer's disease
AD2
The assembly of Aβ and tau proteins into fibrils plays an essential role in mediating the self-propagation and cell-to-cell transmission of pathological amyloid fibrils in Alzheimer's disease. We aim to characterize the heterogeneity of Aβ and Tau amyloid fibrils present in humans, mice and primates and assess the relationships between this diversity and their pathological impacts.
Examples of ongoing projects
- Novel methods for very high molecular assemblies purification form brain tissues and cytosolic extracts (PI. L. Bousset, Cryomet, collaboration with A. A. Arteni, S. Bressanelli, I2BC, Gif sur Yvette, France).
3. Spreading mechanisms in Alzheimer's disease AD3
Self-replication and transcellular spread of tau proteins are responsible for the progressive accumulation of misfolded Tau protein deposits in the brain of Alzheimer's disease patients. Tau spreading correlates with the severity of cognitive decline. Our group characterizes factors mediating and regulating tau spreading as they represent relevant targets for future therapies.
Click on the picture to enlarge
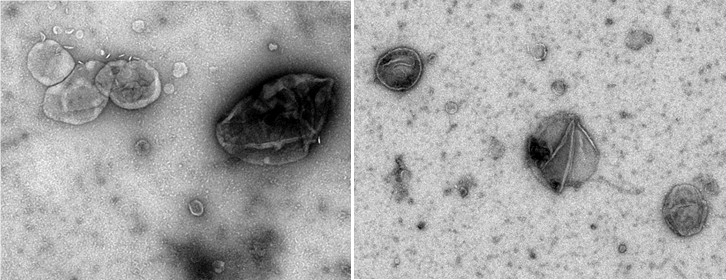
Brain-derived extracellular vesicles visualized by negative-staining electron microscopy (credit M. Kabani)
Examples of ongoing projects
- Molecular profiling of AD brain-derived extracellular Tau (exTau) species -free or associated with lipoproteins, extracellular matrix components or extracellular vesicles- and their role in tau propagation and aggregation (PI. M. Kabani, collaboration with I. Vorberg, DZNE, Germany).
- Assessment of the role of neuronal Heparan-sulfates in extracellular vesicles-mediated Tau pathology spreading (PI. M. Kabani, collaboration with D. Papy Garcia, gly-CRRET, U-PEC, France).
- Assessment of the impact of replication and structural diversification of prions on their cerebral dissemination (PI. M. Dhenain, ANR 2021-2024 (PrionDiff). Collaboration with H. Rezaei, V. Beringue, INRAE, France).
.
Innovative imaging tools for a better understanding of neurodegenerative diseases IM
Neurodegenerative diseases are related to many different "small scale events" (pathological protein accumulation, neuroinflammation, cellular alterations) that lead to large-scale events (tissue loss, neuronal networks alterations, cognitive impairments). Our team develops tools to integrate events occurring at different scales. These new tools require advanced imaging skills combined with artificial intelligence, big data management and high performance computing.
Click on the picture to enlarge
1. 3D microscopic imaging methods based on high performing computing (HPC) and artificial intelligence
IM1
Our group implements image-processing pipelines to perform 3D histology in primates and rodents. 3D-reconstructed brain samples can be analyzed using semi-automatic manual analysis, digital atlas-based analysis (Lebenberg, 2011) or voxel-wise SPM approach without a priori (Vandenberghe, 2018). The method can be used to detect lesions as amyloid plaque related to Alzheimer (Vandenberghe, 2018). New methods are developed to integrate artificial intelligence, massive data management and high performance computing in our analyses. Our methods are mainly developed using in-house software platform BrainVISA (
http://brainvisa.info) and high performance computing (HPC) resources (supercomputer of the TGCC - CEA, Bruyères-le-Châtel).
Click on the picture to enlarge
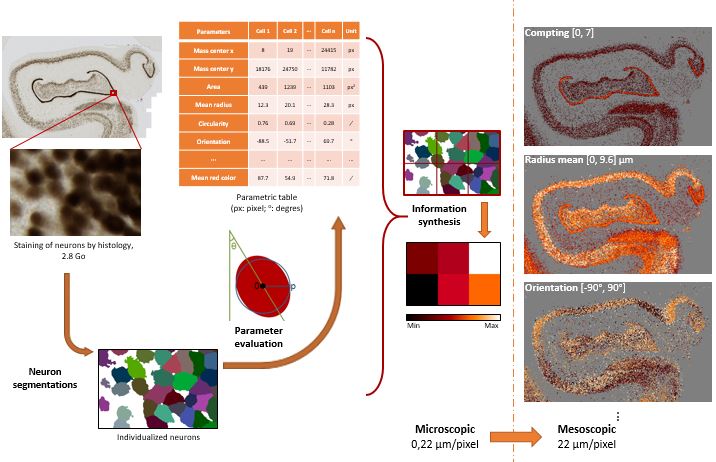
Methods used to evaluate neuronal density and other neuronal parameters based on high performance computing.
Brain sections stained for cells (NeuN antibody) are segmented and various parameters reflecting neuronal characteristics
(e.g. their density, size, orientation, etc…) are calculated.
Based on this analysis, we can produce parametric maps reflecting neuronal states at the level of the whole brain.
These maps can then be compared with other maps (lesion, brain function, etc…).
Examples of ongoing projects
- Neuronal quantification at the level of the whole brain using machine learning algorithm: From random forest to neural networks (PI. T. Delzescaux, Dim Elicit 2020-2023)
- Automatic analysis of 2D histological sections based on 3D digital atlas (PI. T. Delzescaux, Neoxia 2019-2022, CIFRE PhD thesis).
- Computational models for decoding human brain cytoarchitecture (PI. T. Delzescaux, Programme transversal de santé numérique)
- From 3D histology to digital twins of brains – New paradigms to produce realistic mathematical models of cytoarchitecture (PI. T. Delzescaux, collaboration with C. Poupon (NeuroSpin), DAM, Labo anatomie de Tours).
- Small vessel diseases: ultrastructure & microvasculature computational model to refine individual treatment (PI. T. Delzescaux, ANR2021-2024 SUMMIT, collaboration with C. Poupon (NeuroSpin), hôpitaux de Lariboisière, Sainte-Anne, Bretonneau et de Tours)
- New tools for lightsheet imaging analysis (PI. T. Delzescaux, France Relance, Collaboration with J.P. Deslys (CEA-SEPIA) & Imagine Optic (Orsay)).
2. In vivo imaging of functional networks
IM2
Individual cells function in a harmonized way that leads to harmonious brain activity through functional networks. These networks can be assessed by resting state functional MRI and sophisticated image processing tools. Our group studies brain activity with resting state functional imaging. Our group implemented methods to detect neuronal networks in rodents (Celestine, 2020; https://sammba-mri.github.io/; Grandjean, 2020) and primates (Garin, 2021).
Click on the picture to enlarge
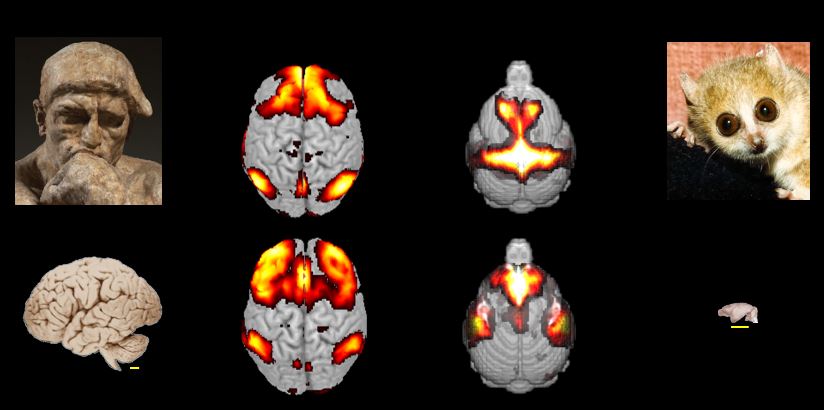
Example of detection of neural networks in humans and in the smallest primate in the world (mouse lemur)
by magnetic resonance imaging.
Examples of ongoing projects
- Digital atlas of mouse lemur brain (PI. J.L. Picq)
- Comparative evaluation of cerebral anatomy and neuronal networks in primates (PI. J.L. Picq, C. Garin)
- Impact of Alzheimer lesions on functional connectivity (PI. M. Dhenain, France Alzheimer, 2021-2023).
- Impact of traumatic brain injury on functional connectivity (PI. M. Dhenain, Service de Santé des Armées).
- StandardRat: A multi-center consensus protocol to enhance functional connectivity specificity in the rat brain (PI. M. Dhenain, Collaboration with J. Grandjean, Radboud University Medical Centre, The Netherlands)
Contributions to Science
Coll
Members of the laboratory associated with these projects MEM
- Luc Bousset (researcher CNRS) -
https://orcid.org/0000-0002-0433-4337
-
Thierry Delzescaux (research director CEA, HDR) - https://orcid.org/0000-0002-6527-7946
- Marc Dhenain (research director , CNRS, DVM, HDR) -
https://orcid.org/0000-0001-8804-4101
- Mehdi Kabani (researcher CNRS, HDR) -https://orcid.org/0000-0001-7440-6394
-
Jean-Luc Picq (researcher Univ. Paris XIII, Professor) -
https://orcid.org/0000-0003-1872-3437
-
Anne- Sophie Hérard (research engineer CEA) - https://orcid.org/0000-0001-8260-9618
- Camille Mabillon (technician CEA, histology)
-
Fanny Petit (technician CEA, histology) -
https://orcid.org/0000- 0003-2757-3434
- Nicolas Souedet (research engineer CEA, high performance computing)
- Marina Célestine (PhD student, Université Paris Saclay)
- Lilian Mehl (France Relance)
- François Plumerault (France Relance)
- Huaqian Wu (PhD student, DIM)
External collaborations COLLE
- Ana -Andreea Arteni, Stéphane Bressanelli, I2BC, Gif sur Yvette, France
- Luc Buée, David Blum - Université de Lille, France
- Alain Buisson - Institut des Neurosciences de Grenoble, France
- Gaël Chételat - Université de Caen, France
- Christos Constantinidis, Université Vanderbilt , USA
- Jean-Philippe Deslys, SEPIA, Fontenay aux Roses, France
- Joanes Grandjean - Radboud University Medical Center, The Net herlands
- Stéphane Haïk, Benoît Delatour - Institut du Cerveau, France
- Han Wei Hou - Nanyang Technological University, Singapour
- Dulce Papy-Garcia - Université Paris Créteil, France
- Fabien Pifferi, Fabienne Aujard - CNRS, Brunoy, France
- Cyril Poupon - Neurospin, Gif sur Yvette, France
- Human Rezaei, Vincent Beringue – INRAE, Jouy en Josas, France
- Stephen Sawiak - Cambridge University, UK
- Elisabeth Traiffort – INSERM, Le Kremlin-Bicêtre, France
- Ina Vorberg – DZNE, Bonn, Allemagne
Industrial partners
Recent grants SUB
- ANR 2020-2024. PrionDiff Impact of replication and structural diversification of prions on their cerebral dissemination
- ANR 2020-2023. SUMMIT: Small vessel diseases: Ultrastructure & microvasculature computational model to refine individual treatment
- France Alzheimer 2022-2023: Impact of specific amyloid strains on Alzheimer’s disease progression: Towards the understanding of the pivotal role of amyloid/Tau interaction on and spreading of Alzheimer’s disease mechanisms on the pathology
- Graduate School LSH – Paris Saclay University 2022. CryoMET. Exploiting flow cytometry methods for ex-vivo purification of extra-large complexes for cryo-EM structure analysis
- FRISBI 2021&2022: Characterization of brain-derived and extracellular vesicles-associated Tau seeds at the origin of Alzheimer’s disease
- Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation-2022-2023. Impact of specific amyloid strains on Alzheimer’s disease progression: Towards the understanding of the pivotal role of amyloid/Tau interaction on and spreading of Alzheimer’s disease mechanisms on the pathology.
- PTC-SN 2021-2022. Modèles computationnels pour le décodage in-vivo de la cytoarchitecture du cortex cérébral humain
- Service santé des Armées . Activation des processus inflammatoires périphériques et centraux à la suite d’un traumatisme crânien : implication des systèmes catécholaminergique et corticotrope
- Région IdF 2019-2022. CARTOBRAIN - Cartographie haute résolution des réseaux de neurones par microscopie optique en recherche préclinique
- DIM Elicit 2020-2023. ENCOMPASS - Evaluation of neuronal counting methods in preclinical studies
- DARI-A10-TGCC. Calcul intensif pour l'analyse par machine learning de données massives 3D de microscopie de cerveaux
- France Relance: image processing developments to improve lightsheet images analysis
Alumni
ALU
Visiting Professor
James Koch, Professeur,Université du Wisconsin, Oshkosh, USA
Post-docs
Salma Bougacha
Nachiket
Matthias Vandesquille
Chrystelle Po
Matthieu Santin
Alexandra Petiet
Oliviero Gobbo
Géraldine Poisnel
Elmahdi Sadouni |
PhD students
Sébastien Piluso
Suzanne Lam
Clément Bouvier
Clément Garin
Zhenzhen You
Yael Balbastre
Michel Vandenberghe
Clémence Dudeffant
Mathieu Bonnet
Charlotte Gary
Anne Bertrand
Olene Dorieux
Nelly Joseph-Mathurin
Audrey Kraska
Jessica Lebenberg
Albertine Dubois
Julien Dauguet | Other students
Alice Fermigier
Emmalaurie Baptiste
Lisa Ciaptacz
Zoé Hanss
Mireilla Peterson
Kelly Herbert
Cecile Cardoso
Maggie Roy
Adrien Pasquier
Myriam Ly
Alexandra Danho
Sang Xuan
Remi Mariani
Guillaume Campeggi
Jean-Luc Lor
Olivier Teboul
Abdelmonem Feki
|
Selected recent publications & thesis
PUB
2022
An evolutionary gap in primate default mode network organization.
Garin C. M., Hori Y., Everling S., Whitlow C. T., Calabro F. J., Luna B., Froesel M., Gacoin M., Ben Hamed S, Dhenain M., Constantinidis C.
Cell Reports. 39, 2, 110669, April 12, 2022: https://doi.org/10.1016/j.celrep.2022.110669.
Macaque neuron instance segmentation only with point annotations based on multiscale fully convolutional regression neural network.
You Z, Jiang M, Shi Z, Shi C, Du S, Liang J, Hérard AS, Jan C, Souedet N, Delzescaux T.
Neural Computing and Applications. 2021. 34, 2925–2938 (2022): https://doi.org/10.1007/s00521-021-06574-7.
An evolutionary gap in primate default mode network organization.
Garin C. M., Hori Y., Everling S., Whitlow C. T., Calabro F. J., Luna B., Froesel M., Gacoin M., Ben Hamed S, Dhenain M., Constantinidis C.
Cell Reports. 39, 2, 110669, April 12, 2022: https://doi.org/10.1016/j.celrep.2022.110669.
Whole brain mapping of glutamate distribution in adult and old primates at 11.7 T.
Garin C. M., Nadkarni N. A., Pépin J., Flament J., Dhenain M.
NeuroImage, 251, 1 May 2022, article. 118984: https://doi.org/10.1016/j.neuroimage.2022.118984.
Neurons with Cat's Eyes: A synthetic strain of α-synuclein fibrils seeding neuronal intranuclear inclusions.
De Giorgi F, Abdul-Shukkoor MB, Kashyrina M, Largitte LA, De Nuccio F, Kauffmann B, Lends A, Laferrière F, Bonhommeau S, Lofrumento DD, Bousset L, Bezard E, Buffeteau T, Loquet A, Ichas F.
Biomolecules. 2022 Mar 11: https://doi.org/10.3390/biom12030436.
Sébastien Piluso (2018-2022) : Automatic segmentation of individual sections of mouse brains by 3D digital atlas. PhD Thesis
2021
Evaluation of automated segmentation algorithms for neurons in macaque cerebral microscopic images.
You, Z., M. Jiang, Z. Shi, X. Ning, C. Shi, S. Du, A. S. Hérard, C. Jan, N. Souedet, Delzescaux T.
Microscopy Research and Technique. 2021: 27 Apr 2021: https://doi.org/2010.1002/jemt.23786.
Alzheimer’s brain inoculation in Aß-plaque bearing mice: synaptic loss is linked to tau seeding and low microglial activity.
Lam S., Boluda S., Hérard A.S., Petit F., Eddarkaoui S., Cambon K., The Brainbank Neuro-CEB Neuropathology Network, Picq J.L., Buée L., Duyckaerts C., Haïk S., Dhenain M.
BioRxiv. 2021. https://doi.org/10.1101/2021.04.06.438654
Resting state functional atlas and cerebral networks in mouse lemur primates at 11.7 Tesla.
Garin C. M., Nadkarni N. A., Landeau B., Chételat G., Picq J-L, Bougacha S., Dhenain M.
NeuroImage, 226, 117589, 2021. https://doi.org/10.1016/j.neuroimage.2020.117589
Suzanne Lam (2017-2021) : "Transmission of Alzheimer pathology in murine and primate models: from proteinopathies to neuronal and cognitive impairments". PhD Thesis
2020
Induction of amyloid-beta deposits from serially transmitted, histologically silent, A-beta seeds issued from human brains.
Herard A.S., Petit F., Gary C., Guillermier M., Boluda S., Garin C. M., French Neuropathology Network, Lam S., Dhenain M.
Acta Neuropathologica Communications. 8, Article number: 205, 2020. https://doi.org/10.1186/s40478-020-01081-7
Modèles primates et innovations thérapeutiques contre les maladies du système nerveux central.
Dhenain M.
La Lettre de l'Académie des Sciences. 2020. n°40, p34. 2020 - https://www.academie-sciences.fr/pdf/lettre/lettre40.pdf.
Effects of chronic masitinib treatment in APPPS1dE9 transgenic mice modeling Alzheimer's disease, 2020.
Li T., Martin E., Abada Y., Boucher C., Cès A., Youssef I., Fenaux G., Forand Y., Legrand A., Nadkarni N., Dhenain M., Hermine O., Dubreuil P., Delarasse C., Delatour B.
Journal of Alzheimer's Disease. 76.4. 1339-1345, 2020. https://doi.org/10.3233/JAD-200466
Sammba-MRI, a library for small animal neuroimaging data processing in Python.
Celestine M.*, Nadkarni N.A.*, Garin C., Bougacha S.*, Dhenain M.
Frontiers in NeuroInformatics. 28 May 2020 | https://doi.org/10.3389/fninf.2020.00024. (These three authors participated equally to the work)
An automated open-source workflow for standards-compliant integration of small animal magnetic resonance imaging data.
Ioanas H-I., Marks M., Garin C. M., Dhenain M., Yanik M. F., Rudin M..
Frontiers in Neuroinformatics. 2020. https://doi.org/10.3389/fninf.2020.00005
Animal functional magnetic resonance imaging: Trends and path toward standardization.
Mandino F., Cerri D. H., Garin C. M., Straathof M., van Tilborg G. A. F., Chakravarty M. M., Dhenain M., Dijkhuizen R. M., Gozzi A., Hess A., Keilholz S. D., Lerch J. P., Ian Shih Y-Y., Grandjean J.
Frontiers in Neuroinformatics. 2020. Vol. 13. Art 78. https://doi.org/10.3389/fninf.2019.00078.
Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis.
Grandjean J., Canella C., Anckaerts C., Ayrancı G, Bougacha S., Bienert T., Buehlmann D., Coletta L., Gallino D., Gass N., Garin C. M. , Nadkarni N. A. , Hübner N., Karatas M., Komaki Y., Kreitz S., Mandino F., Mechling A. E., Sato C., Sauer K., Shah D., Strobelt S., Takata N., Wank I., Wu T., Yahata N., Yun Yeow L., Yee Y., Aoki I. , Chakravarty M. M., Chang W-T., Dhenain M., Von Elverfeldt D., Harsan L. A., Hess A., Jiang T., Keliris G. A., Lerch J. P., Okano H., Rudin M., Sartorius A., Van der Linden A, Verhoye M., Weber-Fahr W., Wenderoth N., Zerbi V., Gozzi A.
NeuroImage. 2020. 205, Article 116278. https://doi.org/10.1016/j.neuroimage.2019.116278
Estimation of COVID-19 cases in France and in different countries: Homogeneisation based on mortality.
Dhenain Marc.
MedRxiv. https://doi.org/10.1101/2020.04.07.20055913
2019
Encephalopathy induced by Alzheimer brain inoculation in a non-human primate.
Gary C., Lam S.*, Herard A.S.*, Koch J.E., Petit F., Gipchtein P., Sawiak S.J., Caillierez R., Eddarkaoui S., Colin M., Aujard F., Deslys J.P., French Neuropathology Network, Brouillet E., Buée L., Comoy E.E., Pifferi F.*, Picq J-L*, Dhenain M.,
Acta Neuropathologica Communications. 2019. 7: 126. https://doi.org/10.1186/s40478-019-0771-x
Promoting healthspan and lifespan with caloric restriction in primates.
Pifferi F., Terrien J., Perret M., Epelbaum J., Blanc S., Picq J.L.*, Dhenain M.*, Aujard F.
Communication Biology, Nature Publishing Group. 2019. 2, 107. 7 https://doi.org/10.1038/s42003-019-0348-z
A 3D population-based brain atlas of the mouse lemur primate with examples of applications in aging studies and comparative anatomy.
Nadkarni N. A, Bougacha S., Garin C., Dhenain M., Picq J.-L.
NeuroImage. 2019. 185. 85-95. https://doi.org/10.1016/j.neuroimage.2018.10.010
Tridimensional mapping of Phox2b expressing neurons in the brainstem of adult Macaca fascicularis and identification of the retrotrapezoid nucleus.
Levy J., Facchinetti P., Jan C., Achour M., Bouvier C., Brunet J.F., Delzescaux T., Giuliano F.
Journal of Comparative Neurology. 2019. May 9. 1-10. https://doi.org/10.1002/cne.24713
Automated individualization of size-varying neurons in 2D microscopic images of macaque brain
You Z, Balbastre Y, Bouvier C., Souedet N., Gipchtein P, Hantraye P, Jan C, Herard A-S, Delzescaux T.
Front Neuroanat. 2019 Dec 17;13:98. https://doi.org/10.3389/fnana.2019.00098
Clément Garin (2015-2019): "Characterization of mouse lemur brain by anatomical, functional and glutamate MRI". PhD Thesis.
OTHERS
Voxel-based statistical analysis of 3D immunostained tissue imaging.
Vandenberghe, M.E., Souedet, N., Herard, A.S., Ayral, A.M., Letronne, F., Balbastre, Y., Sadouni, E., Hantraye, P., Dhenain, M., Frouin, F., Lambert, J.C., Delzescaux, T.
Frontiers in Neuroscience. 2018; 12(article number 754). https://doi.org/10.3389/fnins.2018.00754
A combination of atlas-based and voxel-wise approaches to analyze metabolic changes in autoradiographic data from
Alzheimer's mice.
Lebenberg, J., Herard, A.S., Dubois, A., Dhenain, M., Hantraye, P., Delzescaux, T.
Neuroimage. 2011. 57(4): 1447-1457. https://doi.org/10.1016/j.neuroimage.2011.04.059