Microbiological comparative genomics may be deployed in functional, evolutionary or environmental studies to analyze the genetic content of one or several species of interest. The concept of pangenomics was introduced in the early 2000s with the aim of compiling the entire range of genomic diversity within a species. A pangenome has a "core" genome, comprising the genes present in all of the strains of the species, and a "variable" or "accessory" genome, comprising the genes not present in all of the strains. The variable genome, which represents 5% to 40% of the gene content, has great importance because it provides the species with necessary capacities, for example environmental adaptation or disease resistance.
With the current deluge of new genomics sequences, novel bioinformatics tools are needed to rise to the challenges of analyzing the "Big Data" it creates. It is within this setting that researchers from LABGeM (Genoscope) teamed with colleagues from LaMME (CNRS/University of Évry-Val-d'Essonne mixed research unit) and the Pasteur Institute to develop PPanGGOLiN (Gautreau et al. 2020) a new method for the analysis of the several thousands of prokaryote pangenomes currently available. An originality of PPanGGOLiN is its use of a graph-based approach that depicts not only the entirety of gene families observed in a species but also all genomic colocalization information. Also, PPanGGOLiN comprises an automated graph-structure-based learning method that classifies pangenomes into three subclasses, i.e., "persistent" (gene families present in the majority of genomes), "shell" (present in an intermediate number of genomes) and "cloud" (present in a low number of genomes) genomes. That model proved more efficacious than methods based on gene family frequency thresholds, and furthermore permitted work on lower quality data such as those derived from metagenome¹-assembled genomes (MAGs).
Building upon their pangenome graphs, the LABGeM team then went on to develop a second method called panRGP to predict regions of genome plasticity (RGPs) and their insertion sites or "spots" (Bazin et al. 2020). RGPs usually arise from horizontal gene transfer² and correspond to genomic islands (GIs). Compared to other available GI detection tools, PPanGGOLiN appeared to be the best performing and most rapid method for massive comparative genomics studies.
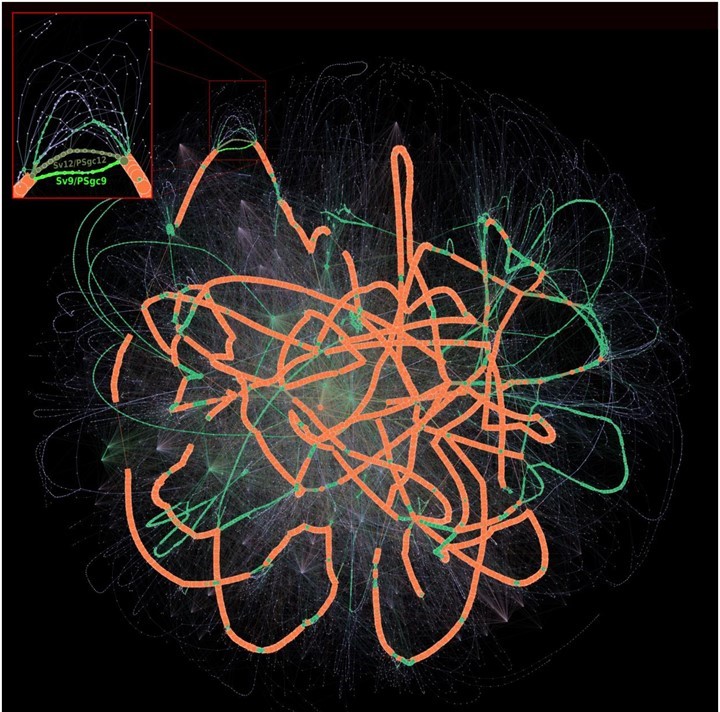
PPanGGOLiN and panRGP are both freely available in the PPanGGOLiN application suite. With this latter, researchers can create and manipulate prokaryote pangenomes from furnished genomic sequences or annotations. These tools are also integrated within the MicroScope platform, with a webpage dedicated to results analyses and explorations.
1 : Metagenomics describes the study of the genetic content of samples collected directly from complex environments (e.g., the intestine, the ocean, soils, etc.) as opposed to samples cultivated in laboratories. Via the direct sequencing of the DNA present in the sample, this approach is able to provide not only a description of the genomic content of the sample but also a glimpse into the functional potential of the environment.
2 : A process by which organisms exchange genetic material independent of lineal descent. Horizontal gene transfer is thus distinguished from vertical gene transfer, as occurs between a parent and offspring.