When an organism is attacked by a pathogen, its immune system mobilizes defense mechanisms via the coordinated and adapted actions of a veritable army of specialized cells. That coordination is essential and enabled by the exchange of signals between cells, either by direct cell-to-cell contact or via soluble immune mediators, including cytokines. Interferon beta (IFN-β) is one of these cytokines. It regulates adaptive immune response to infections and also tumors, notably during radiation therapy. IFN-β production is an essential step in antiviral response and dysregulation of its expression is linked to symptom severity in COVID-19.
Researchers from the Laboratory on Repair and Transcription in Hematopoietic Stem Cells (IRCM) in partnership with the Unit of Human Evolutionary Genetics (Institut Pasteur) have shed light on the regulation of IFN-β expression in the immune system's myeloid cells. Their study, published in PLOS Genetics, suggests that we are not all equal in IFN-β expression by our myeloid cells. In a murine model, the team identified an "enhancer", that is, a distant, non-coding* DNA sequence able to regulate IFN-β expression in the myeloid cells. They then showed that the enhancer was conserved in humans and furthermore that it presented genetic polymorphism, i.e., a punctual difference at a particular position in a gene that coexists in a population. Specifically, an adenine (A) normally situated close to the center of the enhancer (major allele) is substituted by a guanine (G) in about 30% of cases (minor allele). Because of that substitution, the transcription factor C/EBP-β cannot attach to the minor allele's enhancer region, resulting thus in diminished expression and production of IFN-β by the myeloid cells. The team's results illustrate a molecular mechanism linking cellular characteristics with a polymorphism located in a non-coding region of the genome.
To date, the minor allele polymorphism has not been shown to present a risk in human pathologies. Work is underway however to study its importance in recently-described pathophysiological processes involving IFN-β expression, for example COVID-19 or radiation-induced antitumor immune response.
The team's results also led to the filing of a patient application for the use of the polymorphism as a prognostic marker for radiation-induced antitumor immune response. On another front, a hypothesis linking the polymorphism to symptom severity in COVID-19 is being explored as part of the COVIDOGEN project piloted by the CNRGH (CEA-Jacob) and financed by Paris-Saclay University within the framework of its COVID-19 exceptional research program
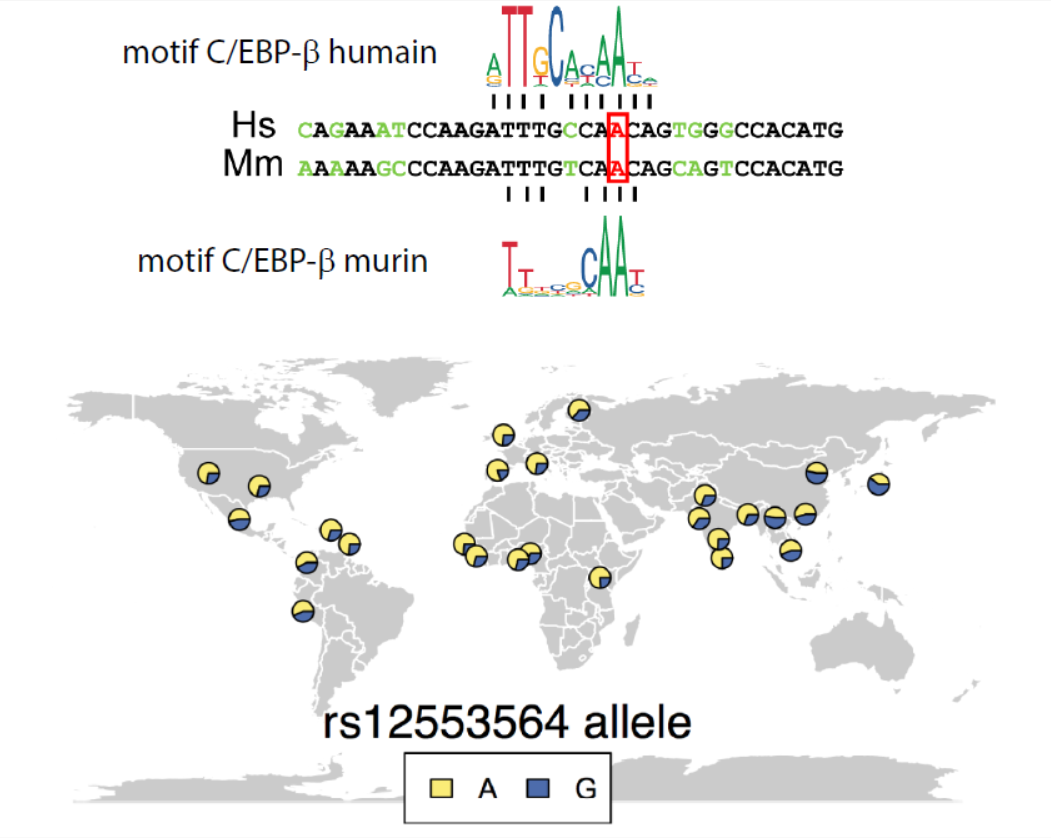
Above: alignment of human and murine DNA sequences at the binding site for the transcription factor C/EBP-β. Below: geographic distribution and proportions of alleles without (yellow) or with (blue) the A to G substitution. (article excerpt © G. Rousselet)
* Non-coding DNA: A DNA sequence that is not translated into a protein.