DIM ELICIT is an Île-de-France network for the development of innovative technologies in life sciences. Coordinated by the Pasteur and Pierre Gilles de Gennes institutes, this network was accredited as a "Domain of Main Interest" (DIM; Domaine d'Intérêt Majeur) by the Île-de-France Regional Council for the 2017–2020 period. Via its calls for projects, DIM ELICIT supports Île-de-France research teams in their efforts to conceive disruptive technologies or disseminate these technologies for applications in life sciences.
In response to the current health crisis, DIM ELICIT launched a project call to fund teams targeting the development of innovative technologies to combat COVID-19 and its causative agent SARS-Cov-2. POSCOVD, a project proposed by SysFate (UMR8030/Genoscope/CEA-Jacob) Team Leader Marco Mendoza, was chosen as a call laureate in July 2020 and presented at the colloquium entitled "Fight SARS-Cov-2: How can innovative technologies for life sciences contribute to the fight against SARS-Cov-2?" and held 20 January 2021.
POSCOVD is aimed at developing a large-scale COVID-19 screening methodology, building particularly upon high throughput sequencing technologies (Oxford Nanopore Sequencing). For the project, Mendoza and his team will:
- evaluate sequencing-based screening sensitivity compared to the currently-employed PCR-based method ;
- estimate the maximum number of different samples that can be analyzed without comprising sensitivity ;
- and explore the methodology's portability via the use of reduced-sized instruments (MinION Mk1C; open-qPCR).
One of the main challenges countries face with COVID-19 is having enough diagnostic tests. Currently, the main diagnostic strategy available for monitoring disease propagation is the use of real-time PCR-based tests, but the effective deployment of these latter is limited by the availability of reagents. Furthermore, the number of tests needed to monitor and control infection propagation must constantly grow with the current tactic of alternating restrictions and lockdowns, and that tendency is only made worse by the recent discovery of more virulent SARS-Cov2 variants.
Within this context, innovative strategies involving massively parallel DNA sequencing methods for population-scale diagnostics are currently being explored not only by international ([1] & [2]) but also by French teams, as illustrated by the efforts of the start-up SeqOne [3]. Although such strategies point to the possibility of analyzing several thousand patient samples per sequencing run, they do necessitate the use of large and costly DNA sequencing facilities piloted by specialized engineers.
The POSCOVD project was conceptualized to address that problem. Specifically, it seeks to develop population-scale massively parallel sequencing-based diagnostics using Oxford Nanopore* technology. This latter's portability and simplicity of use will enable the analysis of large numbers of patient samples even in regions where access to RT-qPCR reagents and/or large DNA sequencing facilities is difficult.
In the protocol under development, each patient's salivary or nasopharyngeal sample is tagged with a molecular "barcode." The samples are grouped thereafter, then amplified and finally sequenced with the Oxford Nanopore technology. A computerized analysis of the data thus obtained will enable the identification of specific infected patients (Figure 1).
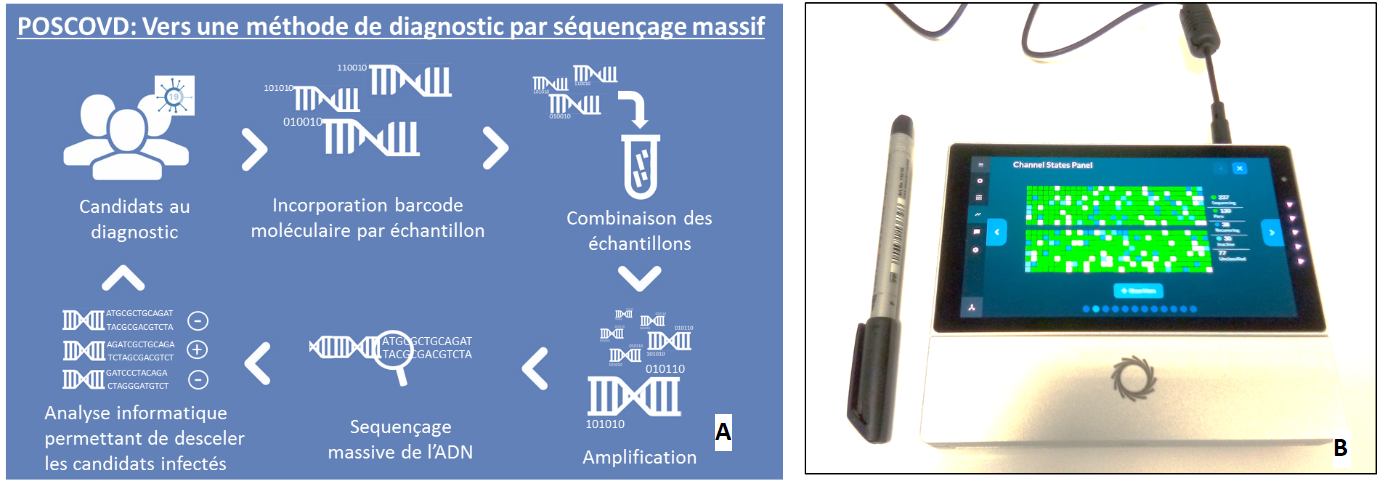
Figure 1 : (Illustration of the main steps in the POSCOVD method. (b) Mk1C DNA sequencing instrument (Oxford Nanopore technology) used in the POSCOVD project. The device's small size (writing pen for scale) will be a veritable advantage for the deployment of the POSCOVD method. Credit Marco Antonio Mendoza/Genoscope
Mendoza and his team are continuing their research to improve the method's sensitivity and specificity and determine the maximum number of samples that can be analyzed simultaneously. They are also taking variant emergence into account in the development of their strategy, as improving the traceability of variants will contribute greatly to monitoring their propagation within populations.
* Oxford Nanopore technology enables DNA molecular sequencing by hydrolyzing the elementary nucleotides (A, T, G, C) and passing them through nanopores. These latter are coupled to electric sensors that identify the specific signature of each constitutive base of the sequenced molecule.
[1] A Massively Parallel COVID-19 Diagnostic Assay for Simultaneous Testing of 19200 Patient Samples; Ayaan Hossain1, Alexander C. Reis2, Sarthok Rahman5, and Howard M. Salis1-4 *
[2] LAMP-Seq: Population-Scale COVID-19 Diagnostics Using a Compressed Barcode Space; Jonathan L. Schmid-Burgk, David Li, David Feldman, Mikołaj Słabicki, Jacob Borrajo, Jonathan Strecker, Brian Cleary, Aviv Regev, Feng Zhang; bioRxv preprint
[3] A French start-up harnesses the power of the latest generation genomic sequencers to dramatically increase COVID 19 testing capacity; 1st May 2020.