
The brown alga Laminaria digitatagrowing on the Brittany coast
Photo : Stéphane Labarre, UMR7139
From an evolutionary point of view, the algae are a heterogeneous group and include organisms from several distinct lineages. The brown algae, or class Phaeophyceae, are part of a group of organisms called the heterokonts (which also includes diatoms and oomycetes) and are only very distantly related to green and red algae (Baldauf, 2003). The vast majority of brown algae occur in marine environments. All members of the group are multicellular, with morphologies ranging from uniseriate branched filaments to complex parenchymatous thalli with multiple cell types, including conducting tissue. The thallus of the kelp Macrocystis pyrifera, for example, can reach up to 70 metres in length. The colour of brown algae is due to the presence of fucoxanthin, a xanthophyll pigment and the principal carbohydrate reserve is laminaran (rather than starch). In addition to cellulose, the cell walls of brown algae contain a variety of other polysaccharides including alginates and fucans.
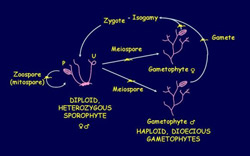
The haploid-diploid life cycle of Ectocarpus siliculosus. The sporophyte and gametophyte are slightly heteromorphic, the gametophyte is dioecious and sex determination is genotypic.
U: unilocular sporangium, P: plurilocular sporangium
Why are brown algae interesting?
- Complex multicellularity - Complex multicellularity has arisen rarely during evolution and the brown algae are one of only five eukaryotic lineages that have evolved this characteristic (the four others being animals, green plants/algae, fungi and red algae). Each of these lineages is thought to have independently evolved the developmental control mechanisms needed for the construction of a complex multicellular organism. In animals and higher plants, the molecular basis of many developmental processes is well understood but surprisingly, considering the fundamental importance of multicellular development, very little is known about this process in the other three lineages with almost nothing being known about the molecular basis of development in the brown and red algae.
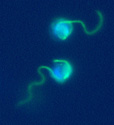
Ectocarpus gametes with their flagella and DNA stained green and blue, respectively. The long undulating flagellum is the anterior one.
(for a scale: the diameter of the lower gamete is 7 µm)
Photo : Delphine Scornet, UMR713
Life cycles and mating types - In addition to being of general interest as complex multicellular organisms, brown algae exhibit several novel features particularly in relation to their life cycles and mating behaviour. Life cycles range from diplontic in Fucus to the nearly isomorphic, haploid-diploid life cycle of Ectocarpus whereas their sexual systems include both dioecious and hermaphrodite states and isogamy, anisogamy or oogamy. Haploid-diploid life cycles like that of Ectocarpus are of particular interest because they imply the existence of genetic control mechanisms that regulate the deployment of the two alternative, independent developmental programmes, influencing development at the level of the whole organism.
Early embryogenesis - Fertilisation is external in brown algae, involving fusion of naked gametes that have been released into the surrounding seawater. This feature has been exploited widely to study many aspects of early development including gamete fusion at fertilisation, establishment of polarity, the involvement of cell walls in early developmental signalling, initiation of the first cell cycle, coupling between the cell cycle and developmental processes and other aspects of development during early embryogenesis (Berger et al., 1994; Bouget et al., 1998; Corellou et al., 2000, 2001; Brownlee et al., 2001). Several cellular phenomena are highly novel in this group; cytokinesis, for example, has been shown to exhibit features typical of both animals and terrestrial plants (Nagasato and Motomura, 2002).
Endosymbiosis and the origin of chloroplasts - The chloroplasts of green plants and red algae are thought to be derived from an ancient cyanobacterium that was engulfed by a eukaryotic host cell. The chloroplasts of heterokonts, such as the brown algae, have a more complicated origin. They have been acquired via a secondary endosymbiotic event in which a chloroplast-containing organism similar to a red alga was itself engulfed by another eukaryote (Yoon et al., 2002). This convoluted history, which has left brown algal chloroplasts with several unusual features such as the four concentric membranes that surround the organelle, involved several phases of transfer of genes to the nucleus. The Ectocarpus genome sequence will allow these events to be traced at the gene level.
Responses to biotic and abiotic stress - The rocky shore habitats of most brown algae are stressful environments in which these organisms are subjected to both biotic aggression from grazers and pathogens and various abiotic stresses including large variations in temperature, immersion, light irradiation and mechanical forces. In consequence, the brown algae represent novel model systems for several aspects of responses to biotic and abiotic stresses including innate immunity (Potin et al., 2002), viral infection (Müller et al., 1998; Delaroque et al., 2001), novel pathosystems (Küpper et al., 2001, 2002; Maier et al., 2000) and osmotic stress (Coelho et al., 2002).
Industrial uses and production of bioactive molecules - In addition to the above fundamental points of interest, it is important to note that seaweeds also represent a marine resource with a wide range of uses in the food, cosmetic, and fertiliser industries and an estimated annual global value of about 4.5x109 euros (McHugh, 2003). They are also attracting increasing attention as a novel source of bioactive molecules, one example being IODUS 40, a formulation that is derived from a storage glucan of Laminaria digitata and which stimulates the natural defence responses of crop plants.

An Ectocarpus sporophyte grown in culture
Photo : Delphine Scornet, UMR7139
Ectocarpus siliculosus: a genetic and genomic model
Brown
algae from the orders Laminariales and Fucales have been studied extensively in
the past but such models are poorly adapted for genomic and genetic approaches
because of their large genomes (ca. 700 and 1100 Mbp, respectively) and because
it is very difficult to complete their life cycles in the laboratory. To
address this problem, a survey of brown algal species was carried out in
Roscoff (Peters et al., 2004a) and this led to the choice of the
filamentous alga, Ectocarpus siliculosus, previously studied over
many years by Dieter Müller at the University of Konstanz, as a model species
for this group. The
choice of Ectocarpus was based on several characteristics including its small size, the fact that the entire life cycle can be completed in Petri dishes in the laboratory (Müller et a.l, 1998), its high fertility and rapid growth (the life cycle can be completed in 2-3 months), the ease with which genetic crosses can be carried out (Peters et al., 2004b) and the relatively small size of the genome. Moreover, the Ectocarpales are closely related to the most economically important brown algal group, the Laminariales (Draisma et al., 2003). Several classical and molecular genetic tools are being developed for Ectocarpusincluding transformation and RNA interference protocols. Mutant lines can be created by UV irradiation and screens for mutants are greatly facilitated by existence of a haploid phase in the life cycle.
Bibliography
-
Baldauf S.L., Roger A.J., Wenk-Siefert I. and Doolittle W.F.
« A kingdom-level phylogeny of eukaryotes based on combined protein data »
Science. (2000) 290 : 972-977. -
Berger, F., Taylor, A. and Brownlee, C.
« Cell fate determination by the cell wall in early Fucus development »
Science. (1994) 263, 1421-1423. -
Bouget F.Y., Berger F. and Brownlee C. .
« Position dependent control of cell fate in the Fucus embryo : role of intercellular communication »
Development.(1998) 125 : 1999-2008. -
Brownlee C., Bouget F.Y. and Corellou F.
« Choosing sides : establishment of polarity in zygotes of fucoid algae »
Semin Cell Dev Biol. (2001) 12 : 345-351 -
Coelho S.M., Taylor A.R., Ryan K.P., Sousa-Pinto I., Brown M.T. and Brownlee C.
« Spatiotemporal patterning of reactive oxygen production and Ca(2+) wave propagation in Fucus rhizoid cells »
Plant Cell. (2002) 14:2369-2381. -
Corellou F., Bisgrove S.R., Kropf D.L., Meijer L., Kloareg B. and Bouget F.Y.
« A S/M DNA replication checkpoint prevents nuclear and cytoplasmic events of cell division including centrosomal axis alignment and inhibits activation of cyclin-dependent kinase-like proteins in fucoid zygotes »
Development.(2000) 127 : 1651-1660. -
Corellou F., Brownlee C., Kloareg B. and Bouget F.Y.
« Cell cycle-dependent control of polarised development by a cyclin-dependent kinase-like protein in the Fucus zygote »
Development. (2001) 128:4383-4392. -
Delaroque, N., Müller, D. G., Bothe, G., Pohl, T., Knippers, R. and Boland, W.
« The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome »
Virology. (2001) 287:112-132. -
Draisma, S. G. A., Peters, A. F. and Fletcher, R. L.
« Evolution and taxonomy in the Phaeophyceae : effects of the molecular age on brown algal systematics »
Norton, T. A. [Ed.] Out of the Past. Collected reviews to celebrate the jubilee of the British Phycological Society. The British Phycological Society, Belfast. (2003) pp. 87-102. -
Maier, I., Parodi, E., Westermeier, R., Muller, D.G.
« Maullinia ectocarpii gen. et sp. nov. (Plasmodiophorea), an intracellular parasite in Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) and other filamentous brown algae »
Protist. (2000) 151, 225-238. -
Müller D.G., Kapp M. and Knippers R.
« Viruses in marine brown algae »
Adv. Virus Res.(1998) 50:49-67. -
Nagasato, C. and Motomura, T.
« Influence of the centrosome in cytokinesis of brown algae : polyspermic zygotes of Scytosiphon lomentaria (Scytosiphonales, Phaeophyceae) »
J. Cell Science (2002) 115 : 2541-2548. -
Peters, A.F., Marie, D., Scornet, D., Kloareg, B. and Cock, J.M.
« Proposal of Ectocarpus siliculosus as a model organism for brown algal genetics and genomics »
Journal of Phycology. (2004) 40, 1079-1088. -
Peters, A.F., Scornet, D., Müller, D.G., Kloareg, B. and Cock, J.M.
« Inheritance of organelles in artificial hybrids of the isogamous multicellular chromist alga Ectocarpus siliculosus (Phaeophyceae) »
Eur. J. Phycol.(2004) 39, 235-242. -
Potin P., Bouarab K., Salaün J.-P., Pohnert G., Kloareg B.
« Biotic interactions of marine algae »
Curr. Op. Plant Biol.(2002) 5 : 308-317. -
McHugh, D. J.
« A guide to the seaweed industry »
FAO Fisheries Technical Paper(2003) No. 441. FAO, Rome, 105 pp. -
Yoon, H.S., Hackett, J.D., Pinto, G., Bhattacharya, D.
« The single, ancient origin of chromist plastids »
Proc. Natl. Acad. Sci., USA.(2002) 99 : 15507-15512