The development of the production management IT system or LIMS (Laboratory Information Management System) began at Genoscope at the start of the 2000s in order to manage sequencing using Sanger technology and in 2008 at the CNG for the management of Illumina technology.
The first high-throughput technology based on technology 454 was incorporated in Genoscope's LIMS in 2007 followed by Illumina technology in 2008.

The initial developments for high-throughput technology were based on what had been done on Sanger technology but it was rapidly clear that there were significant divergences in operational logic.
In parallel, the volume of the data to be processed and hence entered had considerably increased. At practical level, Genoscope's current LIMS version was designed to enable unit entry while the progress of next-generation sequencing (NGS) necessitated being able to process multiple entries.
Adaptation of the LIMS (Genoscope and CNG) to the next-sequencing technologies and the laboratories' new needs for traceability was becoming increasingly difficult. It was therefore decided to develop a common version of next-generation LIMS (NGL) meeting the following specifications:
-
Ensuring the traceability of the Sequencing Laboratory's processes.
-
Adapting to new sequencing technologies on the basis of the progress foreseeable in the next 5 years, to the regular progress of the various associated protocols, to the type of equipment committed, etc.
-
Integrating a scheduling system for the various stages of a sequencing project.
-
Enabling communication with the instruments and, in particular, incorporating any type of automated systems (import / export).
-
Enabling integration of the various types of report (overall, technical, etc.).
-
Ensuring ease of use and providing an intuitive interface.
-
Ensuring real-time system follow-up (incidents, maintenance, etc.).
-
Ensuring reagent batch follow-up (receipt, validation, etc.).
-
Ensuring simplified stock management (management of locations and quantities).
-
Ensuring the specificities of each department (Genoscope and CNG)
Lastly, NGL is not simply a single application. It incorporates various applications able to interact. Each application has its own professional logic but all the applications share the same architecture:
-
NGL-P: project management (data organization and management).
-
NGL-S:
sample management (receipt, validation).
-
NGL-SQ:
feedback management (sequencing, scheduling).
-
NGL-R:
reagent batch management.
-
NGL-BI:
bioinformatics pipeline management.
-
NGL-SUB:
management of submissions in public databanks.
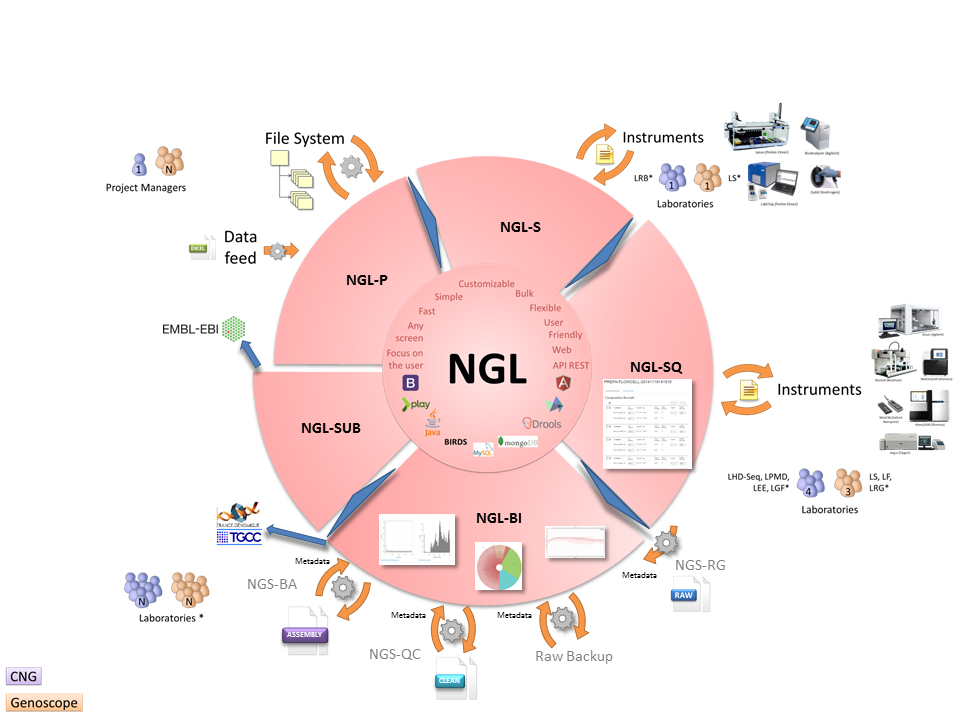
The NGL function in its entirety is thus of primary importance in order to concomitantly ensure experimental traceability, traceability of the bioinformatics data generated and generation of summary reports.
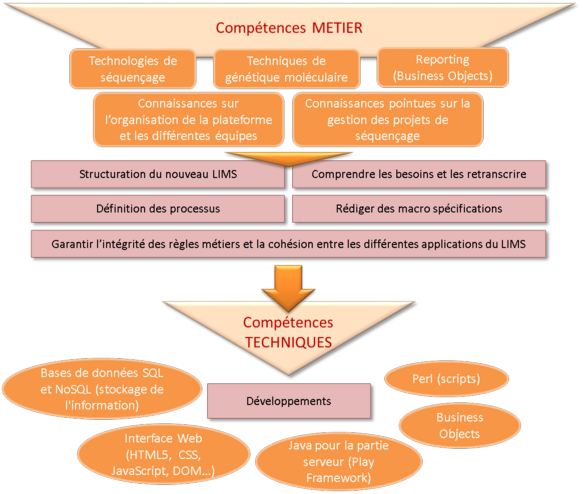
NGL development necessitated professional skills and technical skills.
At professional level, knowledge is indispensable in order to understand the laboratories' requirements and define the specifications:
-
Sequencing technologies (pyrosequencing type 454, Illumina-type synthesis sequencing, single-molecule sequencing using Nanopore technology).
-
Associated molecular genetics techniques.
-
Management of sequencing projects.
-
Reporting.
At technical level, a variety of skills are necessary to implement the NGL:
-
SQL and NoSQL (MongoDB) databases for information storage.
-
Web interface (HTML5, CSS, JavaScript : AngularJS, etc.) for data presentation to the users.
-
Java for the server component (Play Framework).
-
Jboss Drools for management of the professional rules specific to each department.
-
Business Object for report management.
-
Perl for all the scripts.