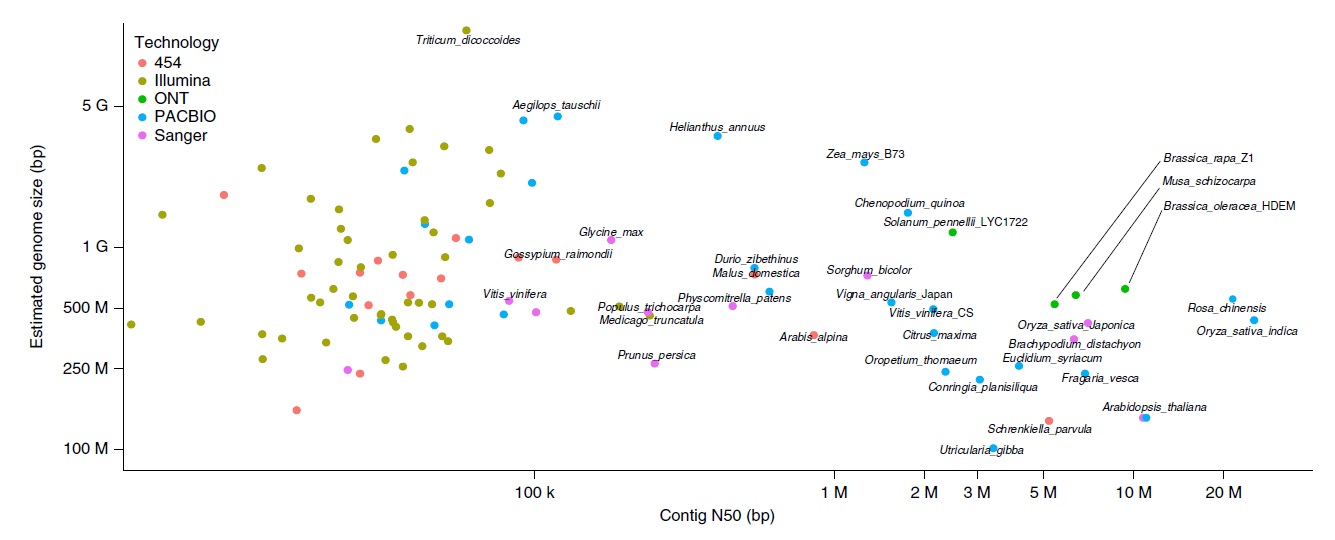
Most sequencing data are currently generated using technology from the company Illumina. Its "short-read" technology can sequence complex genomes, like those of plants, for a relatively low cost, but since its limited to small DNA fragments (100-300 base pairs), it complicates the reconstruction of genomes with numerous repeated sequences. Technologies capable of sequencing long fragments have become available recently, and they do effectively ease the reconstruction of highly repetitive genomes. Nonetheless, even with these "long-read" technologies, reconstructing entire chromosomes had remained out of reach. Until now.
Researchers from Genoscope recently used new genomics technologies to reconstruct the genomes of three species, Brassica rapa (e.g., turnips), Brassica oleracea (e.g., broccoli, cauliflower) and Musa schizocarpa (a wild banana), all notable for their high number of repeated sequences. The genus Brassica has another particularity, specifically a very large intraspecies morphological variability. For example, broccoli, cauliflower, cabbage, German turnips and Brussels sprouts all belong to the species Brassica oleracea. M. schizocarpa is of interest because cultivated bananas are the result of the hybridization of ancestral species; knowledge on the genomes of the latter is vital for characterizing the former. Indeed, the team's objective with the reconstruction of the complete chromosomes of these three species was to lay an essential foundation for future work focused on understanding the morphological differences and the evolutionary history of each variety.
Within a collaborative project involving the CEA, INRA, CIRAD and the CNRS, the researchers assembled readings at the chromosome level for the three species. To get there, the first step was to extract large fragments of DNA and sequence them using a long-read methodology (>50 kb) commercialized by Oxford Nanopore Technology (ONT). However, the assembly of the readings was not sufficient alone for the reconstitution of the chromosomes. Thus, they performed a second step, combining the sequencing data with optical maps produced using the Saphyr system marketed by Bionano Genomics. These optical maps do not furnish information on sequences, but they do furnish information on the organization of the genome at the chromosome level. The resulting genomes count among the most contiguous currently available (see figure below) and have been shared with the scientific community. The combination of these two technologies opens new perspectives for resolving the complexity of the largest plant genomes.