
|
The "Cloaca maxima" project
is investigating the microbial diversity of various domestic waste water
treatment tanks with a particular focus on anaerobic digestion. The process is
widely used in various natural and artificial ecosystems. The process enables
the conversion of complex organic materials into carbon dioxide and methane.
Anaerobic digestion involves a large variety of microorganisms most of which
were unknown when we began the study. |
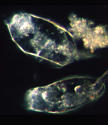
Evry water treatment plant muds.
Photo by P. Ginestet, P.Carmacho (Suez Environnement) |
A conventional metagenomic approach (whole-DNA cloning in the form of large inserts and then systematic sequencing) was implemented in order to study the metabolic capacities of the microorganism flora of the Evry mesophilic anaerobic digester. The first stage was the construction of a representative genome bank (1 million fosmids) and then systematic sequencing of all the extremities of the clones.
 In addition to Archaea, 40 bacterial divisions were detected, showing the great diversity of the microorganisms involved in anaerobic digestion. The metagenomic approach enabled the discovery of 2 candidate bacterial divisions, WWE1 and WWE3 (Pelletier et al., 2008; Guermazi et al., 2008) and the reconstruction of the whole genome of a hitherto uncultured representative of division WWE1 ('Candidatus Cloacamonas acidaminovorans') (Pelletier et al., 2008). Systematic analysis of the assembly (comparison with the nucleic acid and protein resources) and use of functional classification tools enabled analysis of the contigs. A detailed analysis of the 15 scaffolds of more than 1 Mb enabled us to assign a role to the principal contributors (Archaea and bacteria) in the anaerobic trophic chain (cf. figure below).
In addition to Archaea, 40 bacterial divisions were detected, showing the great diversity of the microorganisms involved in anaerobic digestion. The metagenomic approach enabled the discovery of 2 candidate bacterial divisions, WWE1 and WWE3 (Pelletier et al., 2008; Guermazi et al., 2008) and the reconstruction of the whole genome of a hitherto uncultured representative of division WWE1 ('Candidatus Cloacamonas acidaminovorans') (Pelletier et al., 2008). Systematic analysis of the assembly (comparison with the nucleic acid and protein resources) and use of functional classification tools enabled analysis of the contigs. A detailed analysis of the 15 scaffolds of more than 1 Mb enabled us to assign a role to the principal contributors (Archaea and bacteria) in the anaerobic trophic chain (cf. figure below).
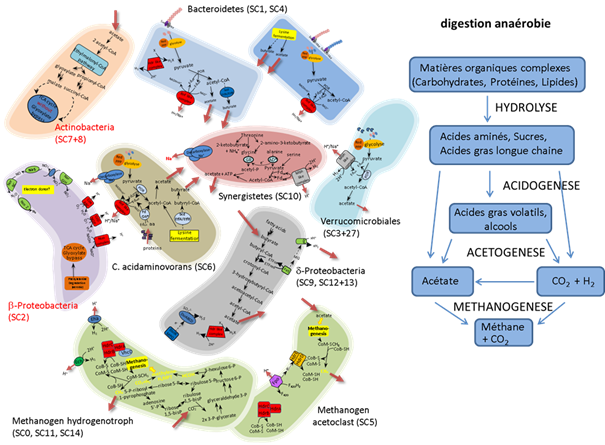
Analysis of the principal scaffolds obtained after assembling the sequences of the Evry mesophilic anaerobic digester and role of the corresponding Archaea and bacteria (Annett Kreimeyer & Nuria Fonknechten).
We also showed that many of the bacterial genomes of the candidate divisions were relatively small (700 to 1500 kb). This has since been confirmed (Brown et al., 2015; Giovannoni et al., 2014; Kantor et al., 2013; Wrighton et al., 2014). In the case of WWE3, analysis of the genomes showed that they all had genes considered essential with the exception of those contributing to biosynthesis of cofactors, hence the difficulty in isolating them.
References:
Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015 Jul 9;523(7559):208-11.
Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014 Aug;8(8):1553-65.
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013 Oct 22;4(5):e00708-13.
Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 2014 Jul;8(7):1452-63.